EFI - Enzyme Similarity Tool
EFI-EST and Cytoscape Tutorials
Network File Download
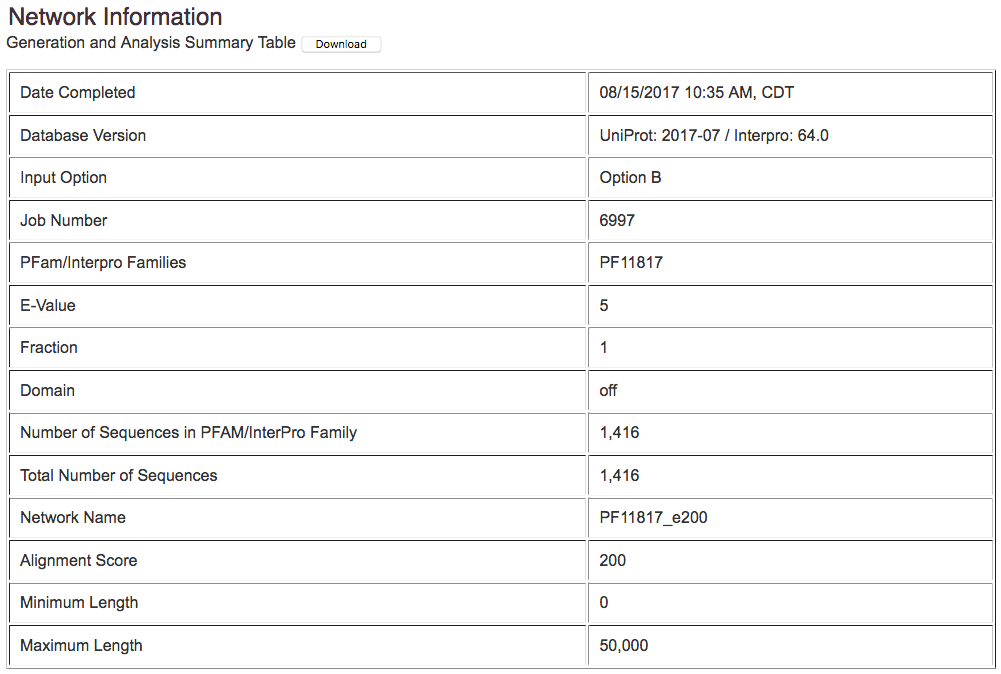
The network file download page includes three tables.
The first displays a summary of the input chosen, and is used for record keeping.
The following tables contain links to download networks, the representative node %ID, the number of nodes, the number of edges, and finally the file size.
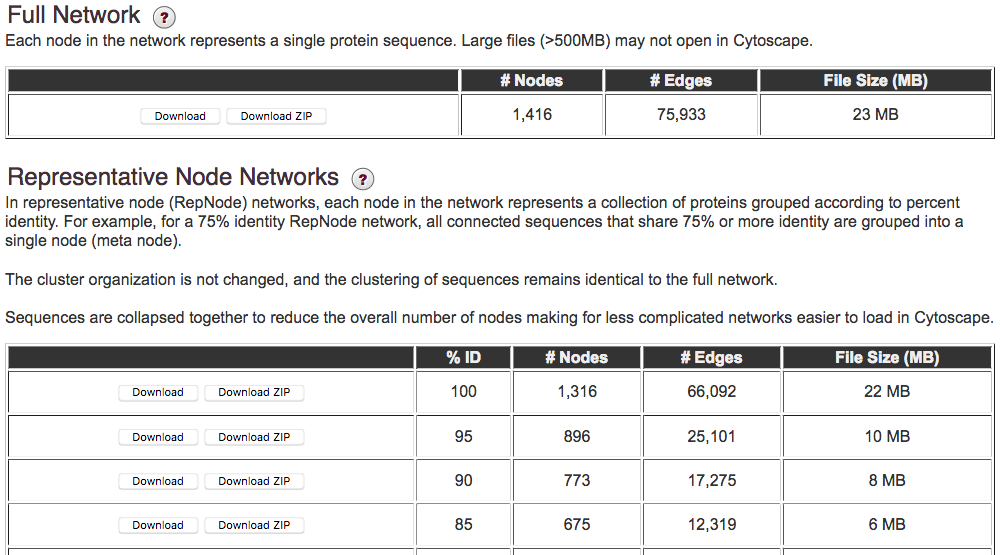
The top table contains the "full" network created at your specified alignment score threshold. By default, this network contains all of the sequences/nodes in your input sequence set. However, this frequently results in very large files (~ 500 MB and greater) that will open and/or run very slowly, or not at all, on most laptop/desktop computers. As a very rough guide, generally Cytoscape networks with a few thousand nodes (protein sequences) and less than ~ 500,000 edges can viewed, although this will depend on your computer. View this "full" network whenever possible, because it will provide access to annotation information for each node in your dataset. Full networks with greater than 10 million edges will not be generated.
In cases where the full network file is too large to open, the bottom table provides the ability to download “representative node” networks. In a representative node (rep node) network, sequences sharing ≥ a specified %ID are grouped into the same node using a program called CD-HIT (4, 5). For example, 90% ID rep node means that each node in the network will contain sequences that share ≥ 90% identity over ANY length of their amino acid sequences. The edges are drawn as done for a full network, except the longest sequence in the rep node is used to determine the alignment score between other rep nodes. For example, if your specified alignment score for the network output was 28, then edges are only drawn between representative nodes where the representative sequences share that alignment score or larger. Rep node networks are automatically calculated at 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, and 100% sequence identity to assure that you will be able to open one or more of the networks on your computer. The number of sequences contained within each rep node as well as the UniProt IDs for those sequences can be viewed in the Cytoscape node attributes panel.
Downloaded files are in the xgmml format and can be imported and viewed in Cytoscape by choosing File → Import → Network and selecting an xgmml file once you have started the Cytoscape program. For more information on using Cytoscape, please see the tutorials here.
Click here to contact us for help, reporting issues, or suggestions.


