EFI - Genome Neighborhood Tool
Viewing and Manipulating a GNN
Cytoscape Requirements: GNN files are opened and viewed in Cytoscape 3.0 (or above) using the Organic layout for the Colored SSN and the Prefuse Force Directed layout for the GNNs.
Opening the colored SSN and both formats of the GNN in a single session of Cytoscape allows fast comparison between the three networks.
Colored SSN: The xgmml file for the colored SSN is opened, visualized using the Organic layout, and analyzed like the SSNs generated with EFI-EST.
GNNs: The Prefuse Force Directed layout places the most connected GNN-clusters at the top of the layout. For the Pfam hub-node format, these are the most commonly occurring Pfam families. For the SSN query cluster hub-node format, these are the SSN clusters that identify the largest number of Pfam families.
For the SSN query cluster hub-node format, the most connected GNN clusters often are those for the input SSN clusters with the fewest number of sequences. This may seem counterintuitive, but the fewer the number of genomes, the higher the co-occurrence for a neighbor. Depending on the minimum co-occurrence frequency selected for displaying Pfam families, those clusters with a small number of sequences will retain many/most of the neighbors and their Pfam families. Therefore, neighbors that are present by "random chance" will be retained, although they are not functionally linked to the query sequences. Many of these will be in the "noise" and removed when the input cluster has a large number of sequences.
For clusters with a large number of sequences, a "large" co-occurrence frequency, e.g., 20%, will eliminate neighbor Pfam families that occur in only a small fraction of the genomes that contain the query sequences. Thus, neighbors that occur by "random chance" and are functionally unlinked to the queries will be excluded from the GNN.
Filtering GNNs: For entire protein families, GNNs generated with a ±10 orf window and a small co-occurrence frequency, e.g., ≤10%, will include a huge amount of information. Therefore, it will be useful, even essential, to filter/simplify the GNNs.
The SSN cluster hub-node format GNN immediately allows the user to focus on the neighbors for individual input SSN clusters. In contrast to filtering a Pfam hub-node GNN, a single cluster is present for each SSN cluster, with spoke-nodes for all of the Pfam families that are identified and satisfy the user-selected co-occurrence frequency. These GNNs will have the same number of clusters as the input SSN.
For Pfam family hub-node GNNs generated with a multiple SSN clusters (in the extreme an entire protein family), the user may want to focus on a single Pfam family to determine whether the input SSN may be "over-fractionated" so multiple clusters find the same genome neighbors. Alternatively, it may be useful to filter this GNN to select one or more specific SSN query spoke-nodes and their directly connected Pfam family hub-nodes and generate a daughter GNN with only those hub- and spoke-nodes; this GNN will contain as many clusters as Pfam families that were identified as neighbors to the SSN cluster.
For SSNs with many clusters, the user can select a specific cluster in the input SSN for more detailed examination, e.g., the SSN cluster hub-node and then its Pfam family spoke-nodes can be selected and a daughter network with the single SSN cluster hub-node can be generated.
The identities of the Pfam families for the neighbors together with their query-neighbor distances and co-occurrence frequencies often is sufficient to distinguish functionally linked neighbors (members of pathways) from functionally unlinked neighbors ("noise").
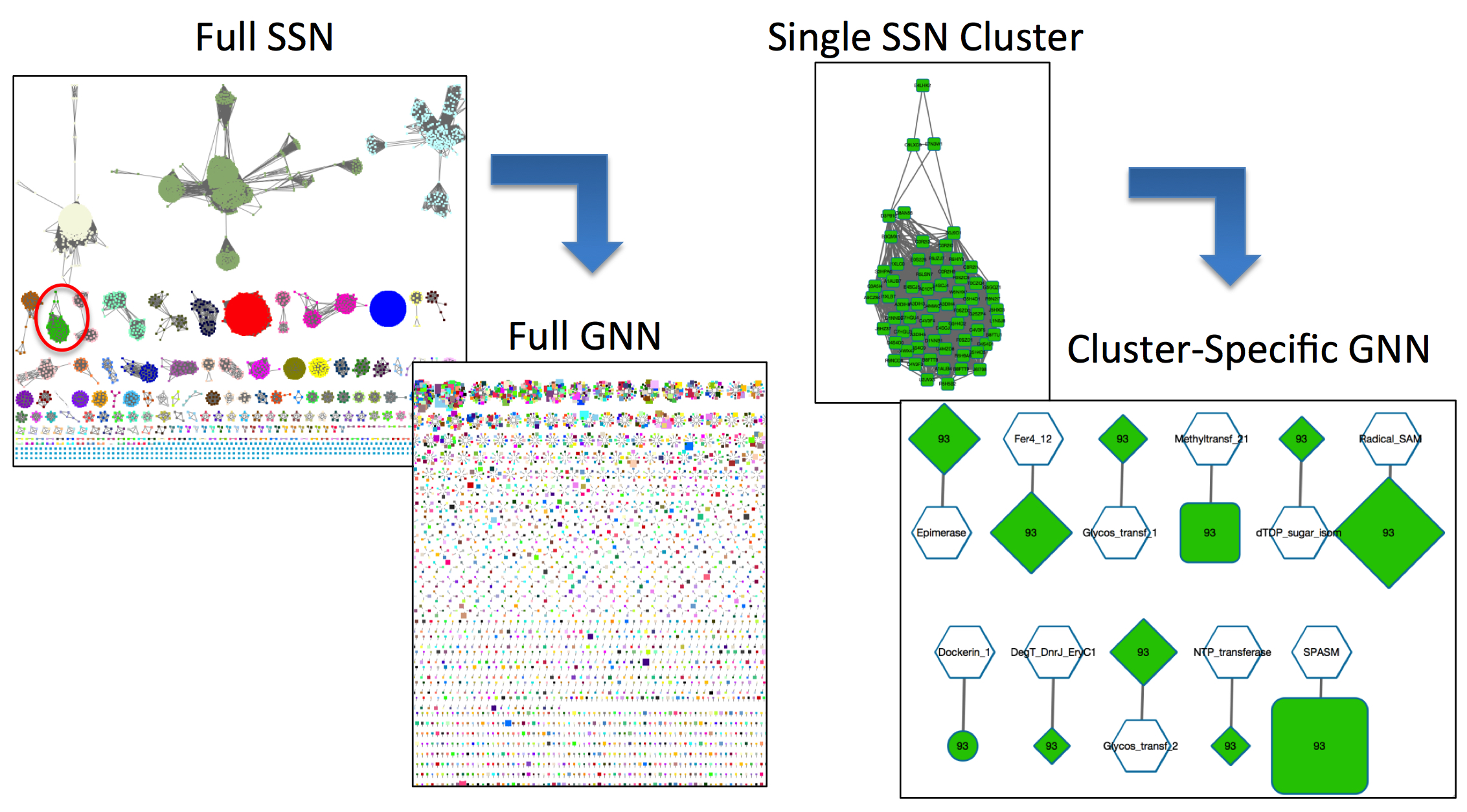
Figure 1. A full GNN prepared with EFI-GNT for the Radical SAM family [left] and a GNN that has been filtered for SSN-cluster 93 [right].
Note that the length of the edge that connects the hub- and spoke-nodes has no significance. Therefore, for crowded GNN-clusters, feel free to click+drag+drop overlapping spoke-nodes until all are visible.
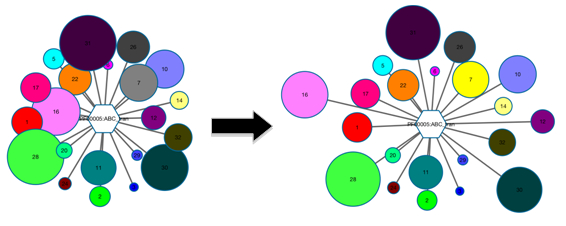
Figure 2. A crowded GNN-cluster can be manipulated to remove overlapping nodes.
Click here to contact us for help, reporting issues, or suggestions.


