EFI - Genome Neighborhood Tool
Result page and file download
EFI-GNT Files and Node Attributes
As described in the previous section, EFI-GNT generates a colored version of the input SSN as well as two formats of genome neighbor networks (GNNs) for download (all three in the xgmml format) and subsequent analysis with Cytoscape.
This section provides a detailed description of the colored SSN and both GNN formats, with emphasis on the node attributes that are provided for the GNNs; these include the query-neighbor distances, co-occurrence frequencies, and the identities of the neighbor’s Pfam family that are used for pathway predictions.
Colored SSN
The colored SSN assists the user in analyzing the GNNs by allowing color-guided association of the cluster nodes in the GNNs with the clusters in the input SSN (Figure 1).
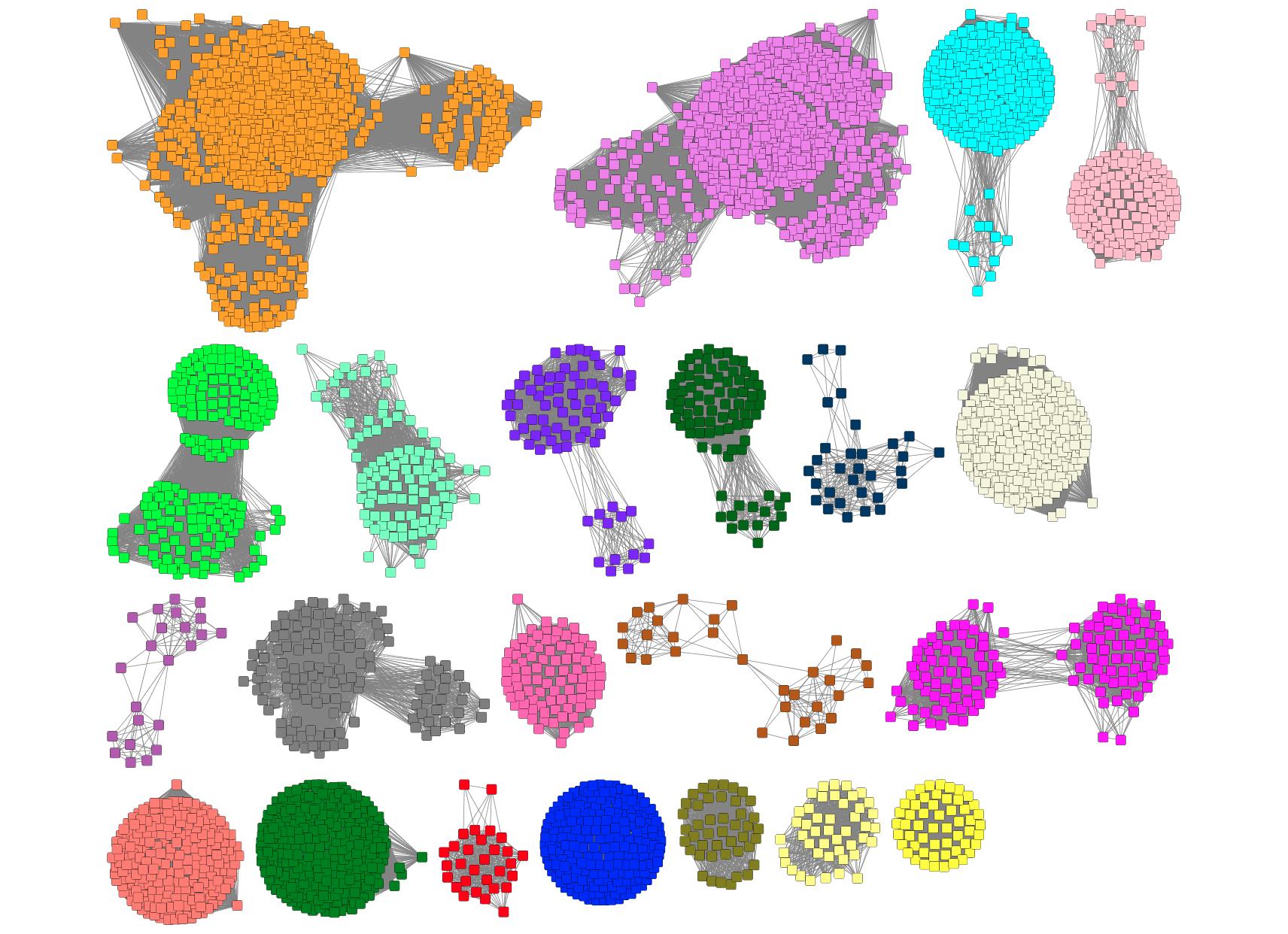
Figure 1. Example of a colored SSN
EFI-GNT assigns a unique number and color to each multi-node cluster in the input SSN. Node attributes are added for the cluster number (Cluster Number) and color (node.fillColor). The clusters are ordered in order of decreasing number of sequences in the clusters (Cluster Sequence Count).
The colored SSN also includes node attributes to indicate whether the sequence has a match in the ENA files (Present in ENA Database?), neighbors in the ENA files (Genome Neighbors in ENA Database?), and the name of the ENA file (ENA Database Genome ID).
In full networks, singletons in the input SSN are excluded from the GNN analysis, while in Rep node networks, singletons with 1 sequence are excluded. Singletons excluded from the GNN analysis will appear in the colored SSN with the Cytoscape default color (cyan). All singletons will not have a cluster number assigned.
Two formats for GNN are generated
EFI-GNT generates the GNN in two formats that provide different query-neighbor perspectives to assist predictions of pathways. The formats differ in the identities of the cluster hub-node (neighbor Pfam family or query SSN cluster) and spoke-nodes (query SSN cluster or Pfam family, respectively).
A GNN contains clusters (hub-node and ≥ 1 spoke‑node) that provide genome neighborhood information (query-neighbor co-occurrence frequencies and distances) for the sequences in the clusters in the input SSN.
In the first GNN format (Figure 2, below), a cluster is present for each query SSN cluster (hub-node, center) that was used to identify genome neighbors (spoke-nodes). This format allows the user to identify functionally linked enzymes, as deduced from genome proximity, that constitute the metabolic pathway in which the sequences in the query SSN cluster participate.
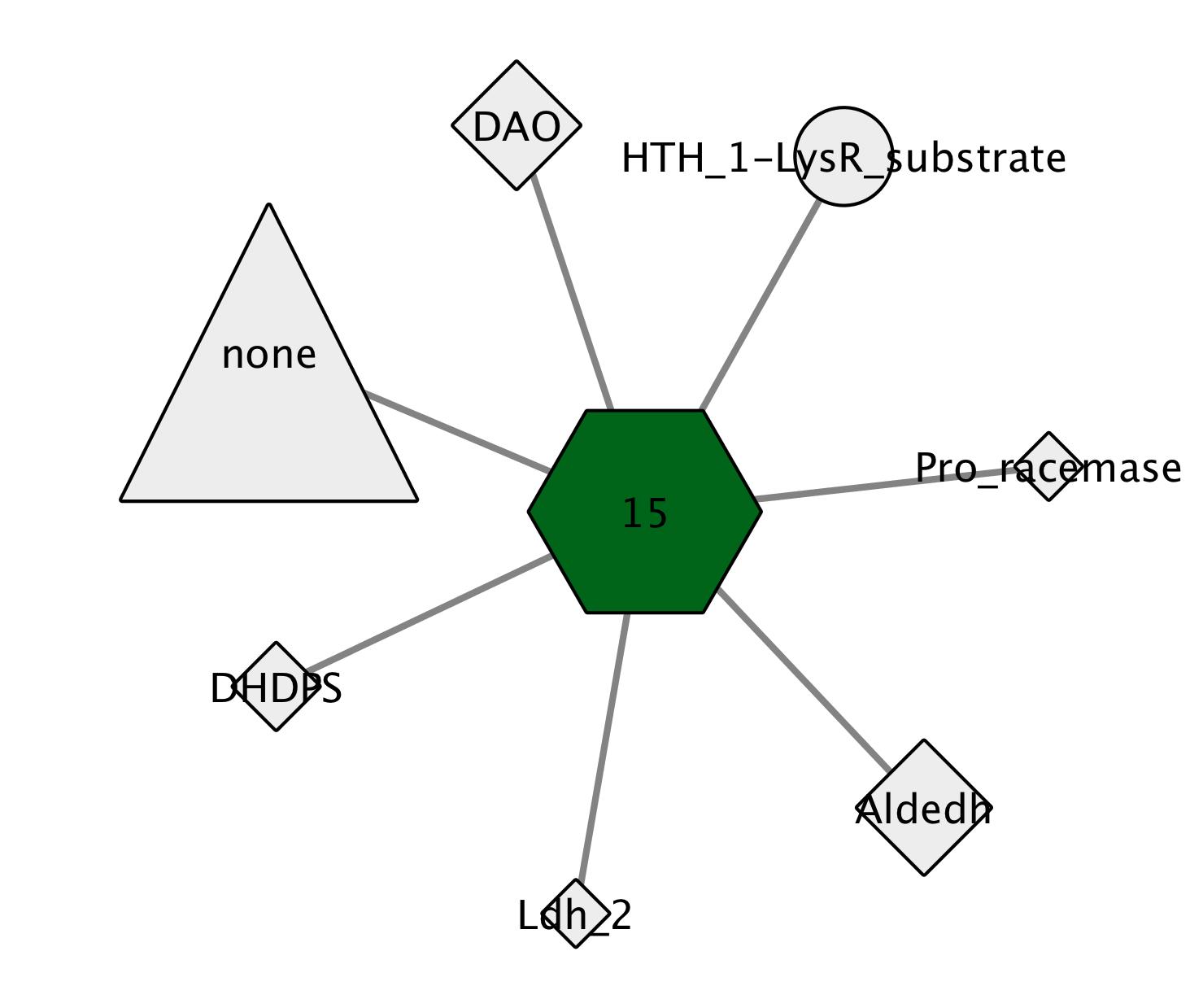
Figure 2. SSN Cluster Hub-Nodes and Pfam Family Spoke-Nodes GNN: The hub-node for each cluster, a hexagon, is the cluster number. The hub-nodes are colored with the unique color assigned for the Colored SSN and labeled with the unique cluster number that was assigned.
In the second GNN format (Figure 3, below), a cluster is present for each Pfam family (hub-node, center) that was identified as a neighbor to queries in the SSN clusters (spoke-nodes). This format allows the user to assess whether queries in multiple SSN clusters are neighbors to members of the same Pfam family and, therefore, may have the same in vitro activities and in vivo metabolic functions.
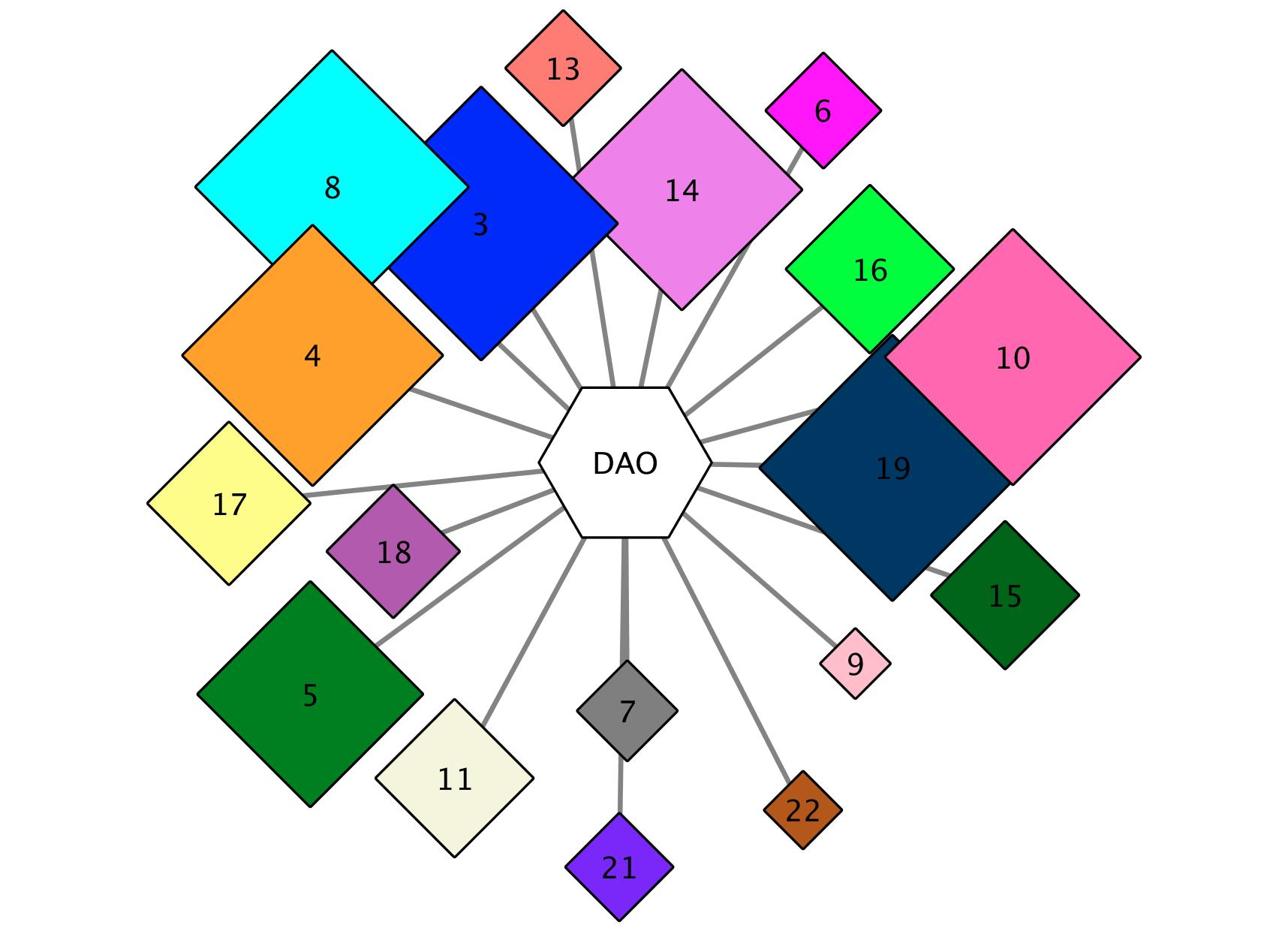
Figure 3. Pfam Family Hub-Nodes and SSN Cluster Spoke-Nodes GNN: The hub-node, a hexagon colored grey, represents the neighbor Pfam family. These are labeled with the Pfam short name for the family (e.g., DAO for PF01266, FAD-dependent oxidoreductase). For multidomain proteins, the label is a composite of the Pfam short names for the component domains (e.g., FGGY_N- FGGY_C for PF00370-PF02782, the N- and C-terminal domains in the FGGY family of carbohydrate kinases).
In both formats, the node attributes for the Pfam family and SSN cluster nodes contain the same information; these provide information about the query-neighbor relationships that can be used to infer functional relationships (co-occurrence frequency and distance) that enable the prediction of in vitro activities and in vivo metabolic pathways.
If the nodes are not automatically colored when the SSN is opened in Cytoscape, the Style Control Panel can be used to color the nodes in the SSN: In the Fill Color property, select "node:node.fillColor" for the Column value and "Passthrough Mapping" for the Mapping Type value.
Additional Files for Download
In addition to the colored SSN, several text files/folders of text files are available for download. These include:
- "UniProt ID-Color-Cluster Number Mapping Table" is a tab-delimited text file with three columns with headers, "UniProt ID", "Cluster Number", and "Cluster Color"; a description of this file and its use was provided in the Color SSN Utility section of the EST Tutorial.
- "UniProt IDs per Cluster" is a folder of files for each cluster that list the UniProt IDs for the sequences in the cluster; a description of the files in this folder and their use was provided in the Color SSN Utility section of the EST Tutorial.
- "FASTA Files per Cluster" is a folder of files for each cluster that contain the FASTA-formatted sequences for the sequences in the cluster; a description of the files in this folder and their use was provided in the Color SSN Utility section of the EST Tutorial.
- "Pfam Neighbor Mapping Tables" is a folder of tab-delimited text files for each neighbor Pfam family that includes the following columns: "Query ID", "Neighbor ID", "Neighbor Pfam", "SSN Query Cluster #", "SSN Query Cluster Color", "Query-Neighbor Distance" (absolute value of distances in ORFs), and "Query-Neighbor Directions" (relative directions of transcription). These files can be used with the Cytoscape BridgeDB App to add these columns as node attributes to the SSN for the neighbor Pfam family so that the neighbors can be located/analyzed in the sequence-function context of the family. The "SSN Query Cluster Color" node attributes can be used with "pass-through mapping" in the Cytoscape Style Color panel to color the nodes in the neighbor Pfam family with the colors in the colored SSN generated by EFI-GNT 1.0, thereby facilitating the determination of whether the neighbors identified in the genome neighborhoods are orthologous. A concatenated file that contains all of the information is also available and may be more convenient for adding node attributes to SSNs for multiple Pfam families.
- "Neighbors without Pfam assigned per Cluster" is a folder of tab-delimited text files for each SSN cluster that lists the accession IDs of neighbors not assigned to Pfam families. These files allow SSNs to be generated with Option D of EFI-EST 2.0 so that protein families not curated by Pfam can be identified. The user should use an alignment score of ~20 to filter the SSN; in most cases, this alignment score will segregate the SSN into protein families.
- "No Matches/No Neighbors File" is a tab-delimited text file with two columns: "UniProt ID" and "No Match/No Neighbor" ("nomatch" or "neighbor"). The same information is included in the colored SSN generated by EFI-GNT 1.0 (vide infra).
Description of node attributes for the various output files is available below and is provided in two formats: a verbose description and a tabular summary.
- Verbose description of node attributes for GNN format #1
- Tabular summary of node attributes for GNN format #1
- Verbose description of node attributes for GNN format #2
- Tabular summary of node attributes for GNN format #2
- Tabular summary of node attributes in colored SSN
Details for format GNN 1: SSN Cluster Hub-Nodes and Pfam Family Spoke-Nodes
In the first GNN format, each hub-node corresponds a cluster present in the SSN (center). The cluster hub-node color and number are identical to those in the colored SSN. Sequences belonging to that cluster were used to retrieve genome neighbors (spoke-nodes; Figure 2). This format allows the user to identify functionally linked enzymes, as deduced from genome proximity, that constitute the metabolic pathway in which the query participates.
A spoke-node, colored grey and represented as a hexagon, is present for each Pfam family that was identified as a neighbor of a query sequence in the cluster represented by the hub-node. These are labeled with the short name for the Pfam family (e.g., Aldedh for PF00171, aldehyde dehydrogenase). For multidomain proteins, the hub-node name is a composite of the short names for the component domains (e.g., HTH_1-LysR for PF00126-PF03466, the N-terminal HTH DNA-binding and C-terminal ligand binding domains of LysR transcriptional regulators). A spoke-node ("none") will be present for neighbors that have not been assigned to a Pfam family.
The size of the spoke-node (node.size) is determined by the value of the Co-occurrence node attribute [decimal value of the ratio of the number of queries (sequences with neighbors) in the cluster that found neighbors in the Pfam family to the number of queriable sequences in the cluster (Queries with Pfam Neighbors/Queriable SSN Sequences); see below]—the larger the co-occurrence frequency of the SSN cluster queries and their genome neighbors, the larger the spoke-node. The value of node.size [calculated as (Co-occurrence * 100)] is used by Cytoscape to draw the node.
The shape of spoke-node (node.shape) is determined by the values of two node attributes for the neighbors that were identified (in the Query-Neighbor Accession node attribute): a triangle if a SwissProt description is available, a square if a Protein Data Bank (PDB) code is available; a diamond if both an EC number and a PDB are available; or a circle if neither an EC number or a PDB code is available. The availability of a SwissProt description and/or a PDB code suggests that the function of the neighbor may be known. The node shape in the node shape node attribute (node.shape) is used by Cytoscape to draw the node. The network can be filtered with the Select Panel to select specific shapes, i.e. different levels of confidence about the functions of the neighbors.
The identities of the neighbors not associated with a Pfam family (no Pfam or "none" family) also are provided in the Neighbor Accession and Query-Neighbor Arrangement node attributes (described in detailed in the descriptions of the node attributes).
Node attributes for each SSN cluster hub-node
cluster#:Pfam#:#Queries with Pfam Neighbors
where- "cluster#" is the cluster# for the query
- "Pfam#" is the spoke-node Pfam family number
- "#Queries with Pfam Neighbors" is the number of queriable sequences in the SSN cluster (# of Sequences in SSN Cluster with Neighbors) for which a neighbor in the spoke-node Pfam family was found.
cluster#:Pfam#:#Pfam Neighbors
where- "cluster#" is the cluster# for the query
- "Pfam#" is the spoke-node Pfam family
- "#Pfam Neighbors" is the number of neighbors in the spoke-node Pfam family found by the queries in hub-node SSN cluster.
cluster#:"Pfam#:average absolute value of distances:median absolute value of distances
where- "cluster#" is the cluster# for the query
- "Pfam#" is the Pfam# for the neighbor
- "average absolute value of distances" is the average of the absolute values of distances between the hub-node queries and spoke-node neighbors
- "median absolute value of distances" is the median absolute value of distances between the hub-node queries and spoke-node neighbors.
cluster#:Pfam#:Co-occurrence:Co-occurrence Ratio
where- "cluster#" is the cluster# for the query
- "Pfam#" is the Pfam# for the neighbor
- "Co-occurrence" is the decimal value of the ratio of the number queries that found neighbors in the Pfam family to the number of queriable sequences in the hub-node SSN query cluster (# of Queries with Pfam Neighbors/# of Sequences in SSN Cluster with Neighbors)
- "Co-occurrence Ratio" is the numerical ratio of the number of queries that found neighbors in the Pfam family to the number of queriable sequences in the hub-node SSN cluster.
If the nodes are not automatically colored when the SSN is opened in Cytoscape, the Style Control Panel can be used to color the nodes in the SSN: In the Fill Color property, select "node:node.fillColor" for the Column value and "Passthrough Mapping" for the Mapping Type value.
Node.size: 70.0 (used by Cytoscape)
Pfam: empty (a spoke-node attribute)
Pfam description: empty (a spoke-node attribute)
# of Queries with Pfam Neighbors: empty (a spoke-node attribute)
# of Pfam Neighbors: empty (a spoke-node attribute)
Query-Accessions: empty (a spoke-node attribute)
Query-Neighbor Accessions: empty (a spoke-node attribute)
Query-Neighbor Arrangement: empty (a spoke-node attribute)
Average Distance: empty (a spoke-node attribute)
Median Distance: empty (a spoke-node attribute)
Co-occurrence: empty (a spoke-node attribute)
Co-occurrence Ratio: empty (a spoke-node attribute)
Node attributes for each Pfam family spoke-node
Pfam#:Query ID:Neighbor ID:EC#:NeighborPDB:ClosestPDB:PDB-E-value:Status
where:- "Query ID" is the query accession ID,
- "Pfam#" is the Pfam# for the neighbor,
- "Neighbor ID" is the neighbor accession ID,
- "EC#" is the E.C. number, if any, assigned to the neighbor in the UniProt database,
- "NeighborPDB is the Protein Databank (PDB) identifier for the neighbor is one is available,
- "ClosestPDB" is the Protein Databank (PDB) identifier for the most similar sequence to the neighbor with a structure in the PDB database,
- "PDB-E-value" is the BLAST e-value for the neighbor-ClosestPDB pair, and
- "Status" (SwissProt/TrEMBL) reports if the in vitro activity of the neighbor has been reviewed by SwissProt.
Pfam#:Query ID:normal/complement:Neighbor ID: noncomplement/complement:Distance
where- "Pfam#" is the Pfam# for the neighbor,
- "Query ID" is the query accession ID,
- "normal/complement" is the direction of transcription of the gene encoding the query (from the ENA sequence file),
- "Neighbor ID" is the neighbor accession ID,
- "normal/complement" is the direction of transcription of the gene encoding the query (from the ENA sequence file), and
- "Distance" is the distance in orfs between the genes encoding the query and neighbor.
# of Sequences in SSN Cluster with Neighbors: empty (a hub-node attribute)
Hub Queries with Pfam Neighbors: empty (a hub-node attribute)
Hub Pfam Neighbors: empty (a hub-node attribute)
Hub Average and Median Distances: empty (a hub-node attribute)
Hub Co-occurrence and Ratio: empty (a hub-node attribute)
Details for GNN format 2: Pfam Family Hub-Nodes and SSN Cluster Spoke-Nodes
In the second GNN format, a cluster is present for each Pfam family (hub-node) that was identified as a neighbor to queries in the SSN clusters (spoke-nodes; Figure 3). This format allows the user to assess whether queries in multiple SSN clusters are neighbors to members of the same Pfam family and, therefore, may have the same in vitro activities and in vivo metabolic functions.
A spoke-node is present for each SSN cluster that identified a member of the Pfam family. The color of the spoke-node is the unique color assigned in the colored SSN; its label is the unique cluster number.
The size of the spoke-node (node.size) is determined by the value of the Co-occurrence node attribute [decimal value of the ratio of the number of queries in the cluster that found neighbors in the Pfam family to the number of queriable sequences in the cluster (# of Queries with Pfam Neighbors/# of Queriable SSN Sequences); see below]—the larger the co-occurrence frequency of the SSN cluster queries and their genome neighbors, the larger the spoke-node. The value of node.size [calculated as (Co-occurrence * 100)] is used by Cytoscape to draw the node.
The shape of spoke-node (node.shape) is determined by the values of two node attributes for the neighbors that were identified (in the Query-Neighbor Accession node attribute): a triangle if a SwissProt description is available, a square if a Protein Data Bank (PDB) code is available; a diamond if both an EC number and a PDB are available; or a circle if neither an EC number or a PDB code is available. The availability of a SwissProt description and/or a PDB code suggests that the function of the neighbor may be known. The node shape in the node shape node attribute (node.shape) is used by Cytoscape to draw the node. The network can be filtered with the Select Panel to select specific shapes, i.e. different levels of confidence about the functions of the neighbors.
The network can be filtered with the Control Select Panel to select specific node shapes, i.e., different levels of confidence about the functions of neighbors.
The identities of the neighbors not associated with a Pfam family are provided in the Neighbor Accession and Query-Neighbor Arrangement node attributes for the Pfam family nodes (described in detailed in the descriptions of the node attributes).
Node attributes for Pfam family hub-nodes
cluster#:Query ID
where- "cluster#" is the cluster# for the query, and
- "Query ID" is the query accession ID.
cluster#:Query ID:Neighbor ID:EC#:NeighborPDB:ClosestPDB:PDB-E-value:Status
where- "cluster#" is the cluster# for the query,
- "Query ID" is the query accession ID,
- "Neighbor ID" is the neighbor accession ID,
- "EC# "is the E.C. number, if any, assigned to the neighbor in the UniProt database,
- "NeighborPDB" is the Protein Databank (PDB) identifier for the neighbor is one is available,
- "ClosestPDB" is the Protein Databank (PDB) identifier for the most similar sequence to the neighbor with a structure in the PDB database,
- "PDB-E-value" is the BLAST e-value for the neighbor-ClosestPDB pair, and
- "Status" (SwissProt/TrEMBL) reports if the in vitro activity of the neighbor has been reviewed by SwissProt.
cluster#:Query ID:normal/complement:Neighbor ID: normal/ complement:Distance
where- "cluster#" is the cluster# for the query,
- "Query ID" is the query accession ID,
- "normal/complement" is the direction of transcription of the gene encoding the query (from the ENA sequence file),
- "Neighbor ID" is the neighbor accession ID,
- "normal/complement" is the direction of transcription of the gene encoding the query (from the ENA sequence file), and
- "Distance" is the distance in orfs between the genes encoding the query and neighbor.
cluster#:Average Distance:Median Distance
where- "cluster#" is the cluster# for the query,
- "average absolute value of distances" is the average distance between the queries and neighbors in the cluster, and
- "median absolute value of distances" is the median distance between the queries and neighbors in the cluster.
cluster#:Co-occurrence:Co-occurrence Ratio
where- "cluster#" is the cluster# for the query,
- "Co-occurrence" is the decimal value of the ratio of the number of queries that found neighbors in the Pfam family to the number of queriable sequences in the cluster (# of Queries with Pfam Neighbors/# of Sequences in SSN Cluster with Neighbors), and
- "Co-occurrence Ratio" is the ratio of the number of queries that found neighbors in the Pfam family to the number of queriable sequences in the cluster.
Average Distance: empty (a spoke-node attribute)
Median Distance: empty (a spoke-node attribute)
Co-occurrence: empty (a spoke-node attribute)
Co-occurrence Ratio: empty (a spoke-node attribute)
Node attributes for SSN cluster spoke-nodes
Query ID:Neighbor ID:EC#:Neighbor PDB:Closest PDB:PDB-E-value:Status
where- "Query ID" is the query UniProt accession ID,
- "Neighbor ID" is the neighbor UniProt accession ID,
- "EC#" is the E.C. number, if any, assigned to the neighbor in the UniProt database,
- "Neighbor PDB" is the Protein Databank (PDB) identifier for the neighbor if one is available,
- "ClosestPDB" is the Protein Databank (PDB) identifier for the most similar sequence to the neighbor with a structure in the PDB database,
- "PDB-E-value" is the BLAST e-value for the neighbor-ClosestPDB pair, and
- "Status" (Reviewed/Unreviewed) reports if the in-vitro activity of the neighbor has been reviewed by SwissProt.
Query ID:normal/complement:Neighbor ID:normal/complement:Distance
where- "Query ID" is the query UniProt accession ID,
- "normal/complement" is the direction of transcription of the gene encoding the query (from the ENA sequence file),
- "Neighbor ID" is the neighbor UniProt accession ID,
- "normal/complement" is the direction of transcription of the gene encoding the query (from the ENA sequence file), and
- "Distance" is the distance in orfs between the genes encoding the query and neighbor.
Pfam description: empty (a hub-node attribute)
Hub Average and Median Distance: empty (a hub-node attribute)
Hub Co-occurrence and Co-occurrence Ratio: empty (a hub-node attribute)
Tabular Summary of Node Attributes
Colored SSN
| Node Attribute | Description - Options A, B, C with FASTA header reading, D |
|---|---|
| Name | Full network - UniProt accession; Rep Node network - UniProt accession for the longest sequence in the representative node (seed sequence for CD-Hit) |
| Shared name | Full network - UniProt accession; Rep Node network - UniProt accession for the longest sequence in the representative node (seed sequence for CD-Hit) |
| Number of IDs in Rep Node1 | Number of UniProt IDs in the representative node |
| List of IDs in Rep Node1 | List of UniProt IDs in the representative node |
| Sequence Source | Options B, C, and D, “USER” if from user-supplied file, “FAMILY” if from user-specified Pfam/InterPro family, “USER+FAMILY” if from both |
| Query IDs | Option C with FASTA header reading and Option D, Input Query ID(s) that identified a UniProt match |
| Other IDs | Option C with FASTA header reading, additional IDs in headers for a FASTA sequence that did not identify a UniProt match (NCBI BLAST files) |
| Cluster Number | Number assigned to cluster, in order of decreasing number of sequences in the clusters (“999999” for singletons) |
| Cluster Sequence Count | Number of sequences in the cluster |
| Node.fillColor | Unique color assigned to cluster, in hexadecimal |
| Present in ENA Database? | “true” if UniProt ID was found in an ENA file (see ENA Database Genome ID); otherwise “false” |
| Genome Neighbors in ENA Database? | “true” if ENA file has sequences for query plus neighbors; “false” if ENA file has no neighbors; “n/a” if not present in ENA database |
| ENA Database Genome ID | ENA file used to obtain genome neighbors |
| Organism | organism genus/genera and species, from UniProt taxonomy.xml |
| Taxonomy ID | NCBI taxonomy identifier(s), from UniProt |
| UniProt Annotation Status | SwissProt - manually annotated; TrEMBL - automatically annotated; from UniProt |
| Description | protein name(s)/annotation(s), from UniProtKB |
| SwissProt Description | protein name(s)/annotation(s), from UniProtKB for SwissProt reviewed entries |
| Sequence Length | number(s) of amino acid residues, from UniProt |
| Gene name | gene name(s) |
| NCBI IDs | RefSeq/GenBank IDs and GI numbers, from UniProt idmapping |
| Superkingdom | domain of life of the organism, from UniProt taxonomy.xml |
| Kingdom | kingdom of the organism, from UniProt taxonomy.xml |
| Phylum | Phylogenetic phylum of the organism, from UniProt taxonomy.xml |
| Class | Phylogenetic class of the organism, from UniProt taxonomy.xml |
| Order | Phylogenetic order of the organism, from UniProt taxonomy.xml |
| Family | Phylogenetic family of the organism, from UniProt taxonomy.xml |
| Genus | Phylogenetic genus of the organism, from UniProt taxonomy.xml |
| Species | Phylogenetic species of the organism, from UniProt taxonomy.xml |
| EC | EC number, from UniProt |
| Pfam | Pfam family, from UniProt |
| IPRO | InterPro family, from UniProt |
| PDB | Protein Data Bank entry, from UniProt |
| BRENDA ID | BRENDA Database ID, from UniProt |
| CAZY Name | Carbohydrate-Active enZYmes (CAZy) family name(s), from UniProt |
| GO Term | Gene Ontology classification(s), from UniProt |
| KEGG ID | KEGG Database ID, from UniProt |
| PATRIC ID | PATRIC Database ID, from UniProt |
| STRING ID | STRING Database ID, from UniProt |
| HMP Body Site | location(s) of organism(s) in/on the body, if human microbiome organism, spreadsheet from HMP |
| HMP Oxygen | oxygen requirement(s), if human microbiome organism, spreadsheet from HMP |
| P01 gDNA | availability of gDNA(s) at EFI Protein Core, in-house |
| Node Attribute | Description - Option C without FASTA header reading |
|---|---|
| Name | zzznnn, where nnn = number of the sequence in FASTA file |
| Shared Name | zzznnn, where nnn = number of the sequence in FASTA file |
| Description | FASTA Header |
| Sequence Length | Length of sequence in FASTA entry |
| Sequence Source | “USER” if from user-supplied file, “FAMILY” if from user-specified Pfam/InterPro family, “USER+FAMILY” if from both |
| Cluster Number | Number assigned to cluster, in order of decreasing number of sequences in the clusters (“999999” for singletons) |
| Cluster Sequence Count | Number of sequences in the cluster |
| Node.fillColor | Unique color assigned to cluster, in hexadecimal |
| Present in ENA Database? | “false” |
| Genome Neighbors in ENA Database? | “n/a” |
| ENA Database Genome ID | none |
SSN Cluster Hub-Nodes and Pfam Family Spoke-Nodes
| Node Attribute | Description - SSN cluster hub-nodes |
|---|---|
| Shared name | Input SSN cluster number |
| Name | Input SSN cluster number |
| Cluster Number | Input SSN cluster number |
| # of Sequences in SSN Cluster | Total number of sequences in SSN cluster |
| # of Sequences in SSN Cluster with Neighbors | Number of sequences in SSN cluster with neighbors (queriable sequences) |
| Hub Queries with Pfam Neighbors | Summary of number of queriable sequences with a neighbor in the Pfam family |
| Hub Pfam Neighbors | Summary of the total # of Pfam neighbors found by the queriable sequences |
| Hub Average and Median Distances | Summary of average and median distances between the query and neighbors in each Pfam family |
| Hub Co-occurrence and Ratio | Summary of the query-neighbor co-occurrence (decimal value) and ratio (fraction) for each Pfam family |
| Node.fillColor | Hexadecimal color for the SSN cluster in the colored SSN, used by Cytoscape |
| Node.shape | "hexagon", used by Cytoscape |
| Node Size | "70.0", used by Cytoscape |
| Node Attribute | Description - Pfam family spoke-nodes |
|---|---|
| Shared name | Pfam family short name |
| Name | Pfam family short name |
| SSN Cluster Number | SSN Cluster that found neighbors in the Pfam family |
| Pfam | Pfam family number (PFnnnnn) |
| Pfam description | Pfam family description |
| # of Queries with Pfam Neighbors | Number of queriable sequences with a neighbor in the Pfam family |
| # of Pfam Neighbors | Number of Pfam neighbors found by the queriable sequences |
| Query-Accessions | List of SSN cluster queries that found neighbors in the Pfam family |
| Query-Neighbor Accessions | Information about query-neighbor pairs in the Pfam family |
| Query-Neighbor Arrangement | Genome context information for the query-neighbor pairs in the Pfam family |
| Average Distance | Average distance (in ORFs) between the SSN cluster queries and Pfam neighbors |
| Median Distance | Median distance (in ORFs) between the SSN cluster queries and Pfam neighbors |
| Co-occurrence | Decimal value of ratio of queries that found neighbors to queriable sequences |
| Co-occurrence Ratio | Ratio of queries that found neighbors to queriable sequences |
| Node.fillColor | #EEEEEE, grey in hexadecimal, used by Cytoscape |
| Node.shape | "ellipse", "diamond", or "square"; explained in on-line tutorial, used by Cytoscape |
| Node.size | Co-occurrence * 100, used by Cytoscape |
Pfam Family Hub-Nodes and SSN Cluster Spoke-Nodes
| Node Attribute | Description - Pfam family hub-nodes |
|---|---|
| Shared name | Pfam family short name |
| Name | Pfam family short name |
| Pfam | Pfam family number (PFnnnnn) |
| Pfam description | Pfam family description |
| # of Sequences in SSN Cluster | Total number of sequences in SSN cluster |
| # of Sequences in SSN Cluster with Neighbors | Number of sequences in SSN cluster with neighbors (queriable sequences) |
| # of Queries with Pfam Neighbors | Number of queriable sequences with a neighbor in the Pfam family |
| # of Pfam Neighbors | Number of Pfam neighbors found by the queriable sequences |
| Query-Neighbor Accessions | Information about query-neighbor pairs in the Pfam family |
| Query-Neighbor Arrangement | Genome context information for the query-neighbor pairs in the Pfam family |
| Hub Average and Median Distances | Summary of average and median distances between the query and neighbors |
| Hub Co-occurrence and Ratio | Summary of the query-Pfam family co-occurrence (decimal value) and ratio (fraction) |
| Node.fillColor | “#FFFFFF”, white in hexadecimal, used by Cytoscape |
| Node.shape | “hexagon”, used by Cytoscape |
| Node.size | “70.0”, used by Cytoscape |
| Node Attribute | Description - SSN cluster spoke-nodes |
|---|---|
| Shared name | Input SSN cluster number |
| Name | Input SSN cluster number |
| Cluster Number | Input SSN cluster number |
| # of Sequences in SSN Cluster | Total number of sequences in SSN cluster |
| # of Sequences in SSN Cluster with Neighbors | Number of sequences in SSN cluster with neighbors (queriable sequences) |
| # of Queries with Pfam Neighbors | Number of queriable sequences with a neighbor in the Pfam family |
| # of Pfam Neighbors | Number of Pfam neighbors found by the queriable sequences |
| Query-Accessions | List of queries in each SSN cluster that found neigbhors in the Pfam family |
| Query-Neighbor Accessions | Information about query-neighbor pairs in the Pfam family |
| Query-Neighbor Arrangement | Genome context information for the query-neighbor pairs in the Pfam family |
| Average Distance | Average distance (in ORFs) between the SSN cluster queries and Pfam neighbors |
| Median Distance | Median distance (in ORFs) between the SSN cluster queries and Pfam neighbors |
| Co-occurrence | Decimal value of ratio of queries that found neighbors to queriable sequences |
| Co-occurrence Ratio | Ratio of queries that found neighbors to queriable sequences |
| Node.fillColor | Hexadecimal color for the SSN cluster in the colored SSN, used by Cytoscape |
| Node.shape | “ellipse”, “diamond”, or “square”; explained in on-line tutorial, used by Cytoscape |
| Node.size | Co-occurrence * 100, used by Cytoscape |
Click here to contact us for help, reporting issues, or suggestions.


