EFI - Genome Neighborhood Tool
GNN Generation Process
A genome neighborhood network (GNN) is generated in two steps.
Step 1: SSN upload
A sequence similarity network (SSN) from EFI-EST partitioned into "isofunctional" clusters with an appropriate alignment score, using either the Analyze step of EFI-EST or by filtering with Cytoscape, is the input.
Acceptable SSNs are generated for an entire Pfam and/or InterPro protein family (from Option B of EFI-EST), a focused region of a family (from Option A of EFI-EST), a set of protein sequence that can be identified from FASTA headers (from option C of EFI-EST with header reading) or a list of recognizable UniProt and/or NCBI IDs (from option D of EFI-EST). An SSN manually modified within Cytoscape that originated from any acceptable EFI-EST Option is also acceptable. SSNs that have been colored using the "Color SSN Utility" of EFI-EST and that originated from any EST option are also acceptable.
The .xgmml file for the SSN is the input for EFI-GNT and is uploaded by the user on the Start page. EFI-GNT recognizes the clusters in the SSN and extracts the UniProt accession IDs for the sequences in each cluster. Each cluster is assigned a unique cluster number, and the nodes for the sequences in each cluster are assigned a unique color. For "full networks", singletons in the SSN are excluded from the analysis, although they will be present in the colored SSN that is provided by EFI-GNT (with the default Cytoscape color, cyan). For Rep node networks, singletons containing a single sequence will be excluded from the analysis. EFI-GNT provides a numbered and colored version of the SSN (as an xgmml file) to assist the user in analyzing the GNNs. The colored SSN and GNNs share identical coloring and number of clusters for easy cross-referencing.
Step 2: Gathering neighboring information
The sequences constituting each cluster are identified. EFI-GNT then queries the STD (annotated assembled sequences), CON (high level constructed sequences), and WGS (whole genome shotgun sequencing with intermediate level of assembly) sequence files for bacterial (prokaryotic and archaeal, PRO), fungal (FUN), and environmental (ENV) entries in the European Nucleotide Archive (ENA) database for the neighbors of each sequence in a cluster.
The default window for identifying neighbors is ± 10 orfs from the query sequence—the user can select a smaller/larger window on the Start page (from ±3 to ± 20 orfs). As the size of the window decreases, the signal-to-noise in the GNN increases, although smaller windows may miss functionally linked neighbors.
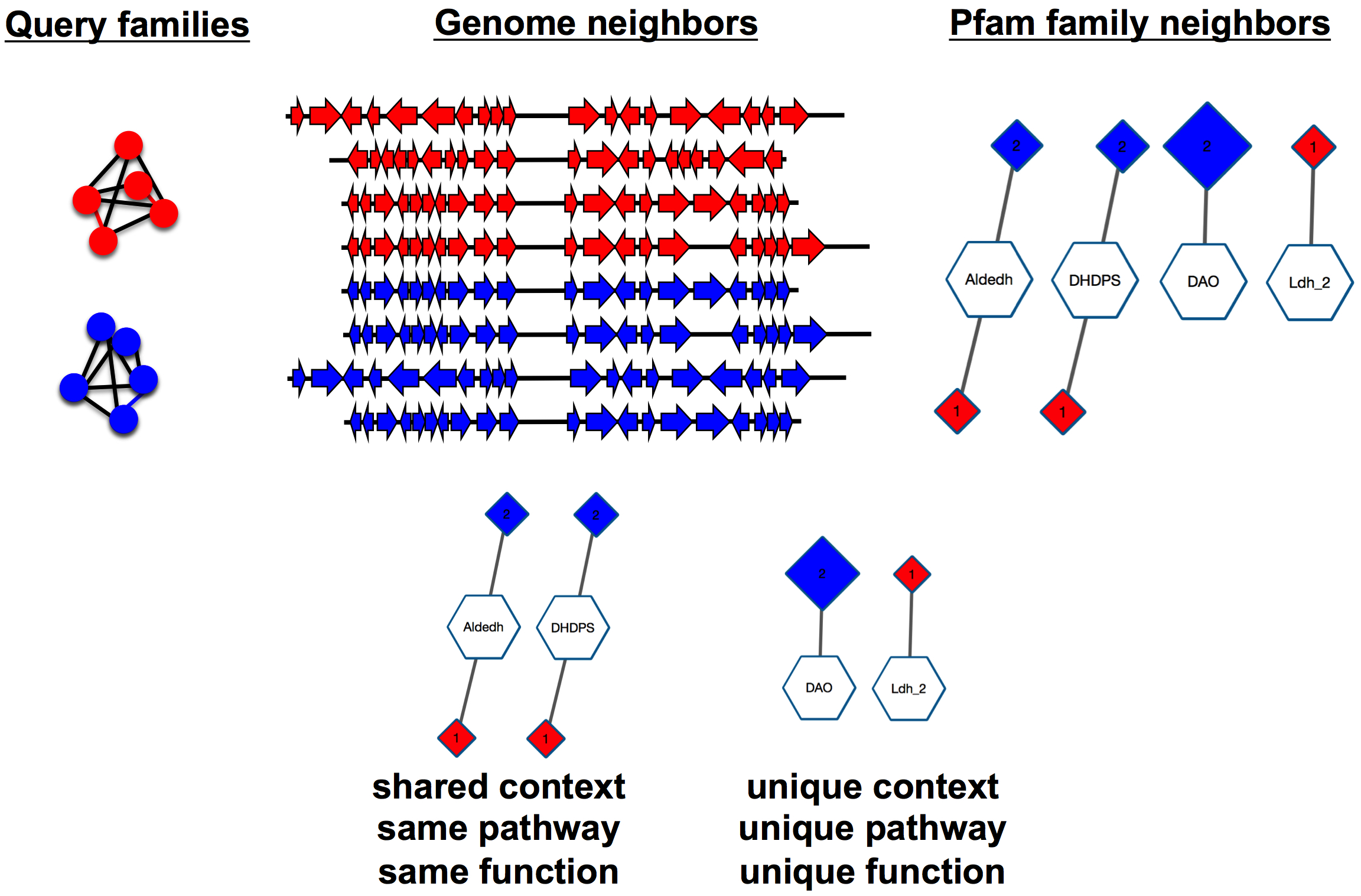
The 20 neighbors collected with the ± 10 orf default (or 2N neighbors collected with a user-specified ±N orf window) constitute the genome neighborhood for the query. If a neighbor is an annotated RNA (rRNA or tRNA), it is discarded although its "place" is included in the ± 10 orf count. Each of the protein neighbors is then associated with a Pfam family using annotations provided by the UniProt and InterPro databases.
For each query-neighbor pair, the EFI-GNT collects the distance (in orfs), genome start/stop coordinates for the query and neighbor, and the direction of transcription for the query and neighbor (normal or complement strand).
Multidomain proteins: If a neighbor is a multidomain protein, i.e., containing multiple domains defined by Pfam, EFI-GNT reports that the neighbor is multidomain by providing a hyphenated list of the Pfam family names for the domains as the name of the GNN node (hub or spoke) for the neighbor Pfam family; EFI-GNT also provides hyphenated lists of all of the Pfam family names and numbers as node attributes for the Pfam family node (name, shared name, Pfam, and Pfam description).
None, or Neighbors not in Pfam: If a neighbor is not associated with a Pfam family (~20% of the proteins in UniProt are not assigned to a Pfam family), it is assigned to the "no Pfam" family. The "no Pfam" family is included in the GNNs (as clusters labeled "none"). A file containing the UniProt IDs of the "no Pfam" neighbors is available for download so that a SSN for the "none" family can be generated using Option D of EFI-EST, thereby allowing these to be placed into families that have not (yet) been curated by Pfam.
Genomes not in ENA files: Bacterial (prokaryotes and archaea) and fungal genomes often are organized in operons and/or gene clusters that encode pathways, so these are mined for genome neighborhoods. Because EFI-GNT only queries ENA files for these organisms, some queries in the input SSN, e.g., encoded by plant and mammalian genomes, will not find matches in these files. In addition, because of the nature of the release schedule of the UniProt protein sequence files and ENA nucleotide sequence files, some bacterial and fungal entries in UniProt may not have entries in the ENA database used by EFI-GNT so no matches will be found. A file containing the UniProt IDs for queries with no matches in the ENA files is available for download; in addition, these are identified in the colored SSN with the Present in ENA Database? node attribute.
Queries with no or incomplete genome context: Not all sequences in the query SSN will identify 20 neighbors with the ± 10 orf default window, excluding the RNAs, (or 2N neighbors with a user-specified ±N orf window). Depending on the organism and/or the type of sequencing project that contributed the ENA file in which the query sequence is located, a smaller number of neighbors, sometimes no neighbors, may be found if the query is close to/at the end of a contig or linear chromosome. A file containing the UniProt IDs for queries with no neighbors in the ENA files is available for download; in addition, these are identified in the colored SSN with the Genome Neighbors in ENA Database? node attribute.
Result page and file download
Several files are generated for download, including the colored version of the input SSN (details in the next section), the two formats of the GNN (details in the next section), and various text/spreadsheet files that can be used for multiple sequence alignments and custom node attributes for analyses of the neighbors in the SSNs for their families.
Click here to contact us for help, reporting issues, or suggestions.


