EFI - Chemically-Guided Functional Profiling
Marker Quantification/Metagenome Abundance
When the marker identification step is completed, EFI-CGFP sends an e-mail to the user. In the "Previous Jobs" tab, the job name will be changed to a green-colored link to the "Markers Computation Results" page.
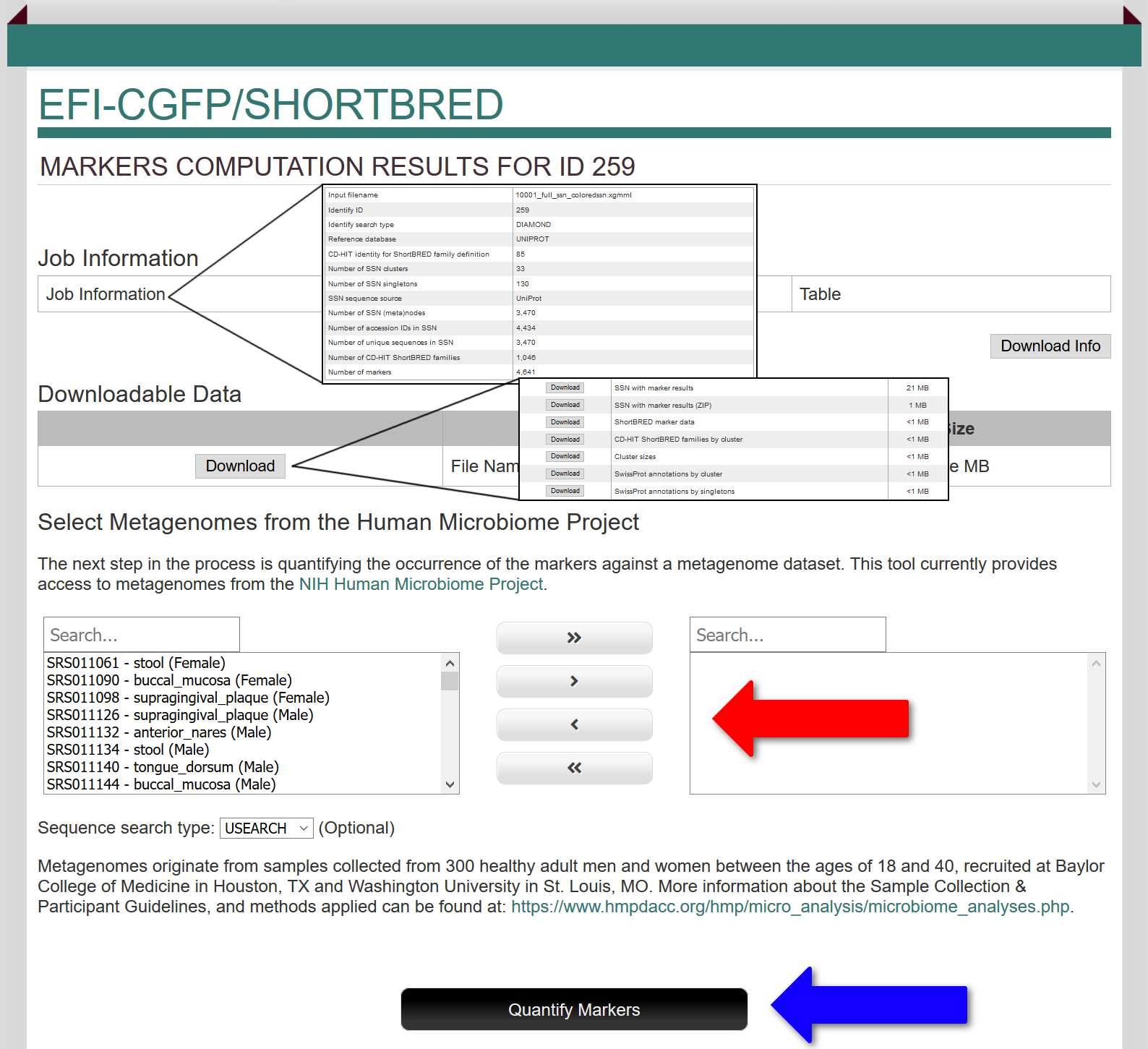
A table summarizing the Job Information is provided at the top of the page. The information includes the parameters used for marker identification as well job statistics about the input SSN, e.g., number of clusters, number of singletons, number of (meta)nodes, total number of accession IDs, number of unique sequences, number of CD-HIT85 ShortBRED families, and number of markers. This information can be downloaded as a text file.
This page provides seven files that can be downloaded: 1) the input Colored SSN to which node attributes have been added containing the type and number of markers that were generated ("SSN with marker results"); 2) the zipped file for the same SSN; 3) a tab-delimited text file ("Marker data") that provides the identities of and information about the markers that were generated; 4) a tab-delimited text file that identifies the seed sequences for the CD-HIT 85 ShortBRED families that were generated and the sequences that are contained in each cluster ("CD-HIT mapping file"); 5) the number of sequences in the SSN clusters (from the input colored SSN); 6) the SwissProt annotations associated clusters (and the accession ID for the SwissProt description from the input colored SSN); and 7) the SwissProt annotations associated with singletons.
The identify job SSN adds four additional node attributes to the input SSN: 1) "Seed Sequence" with the accession ID if the (meta)node is/contains the seed (longest) sequence for a CD-HIT 85 ShortBRED family; 2) "Seed Sequence Cluster(s)" with the accession ID of the seed sequence for the ShortBRED family with which the accession ID(s) was(were) grouped; 3) "Marker Types" for the markers identified in the seed sequences (True, Quasi, or Junction; see the ShortBRED article for details); and 4) "Number of Markers" with the number of markers identified in the seed sequence.
This page also enables the user to select the Human Microbiome Project (HMP) metagenome datasets from a library of 380 datasets for healthy adult men and women from six body sites [stool, buccal mucosa (lining of cheek and mouth), supragingival plaque (tooth plaque), anterior nares (nasal cavity), tongue dorsum (surface), and posterior fornix (vagina)]. Subsets of the library can be selected using the "Search" boxes.
We plan to add additional libraries of metagenome datasets, e.g., intestinal bowel disease metagenomes and transcriptomes from HMP2.
After the datasets are selected (red arrow; transferred from the left panel to the right panel), metagenome abundance is initiated with the "Quantify Marker" button (blue arrow). On the "Previous Jobs" tab, the quantify job will appear with a list of the selected metagenomes. Quantitiation of metagenome may require >24 hrs for execution.
The "Existing Quantify Jobs" button is provided to access the pages for the metagenome quantification jobs/results generated with the markers.
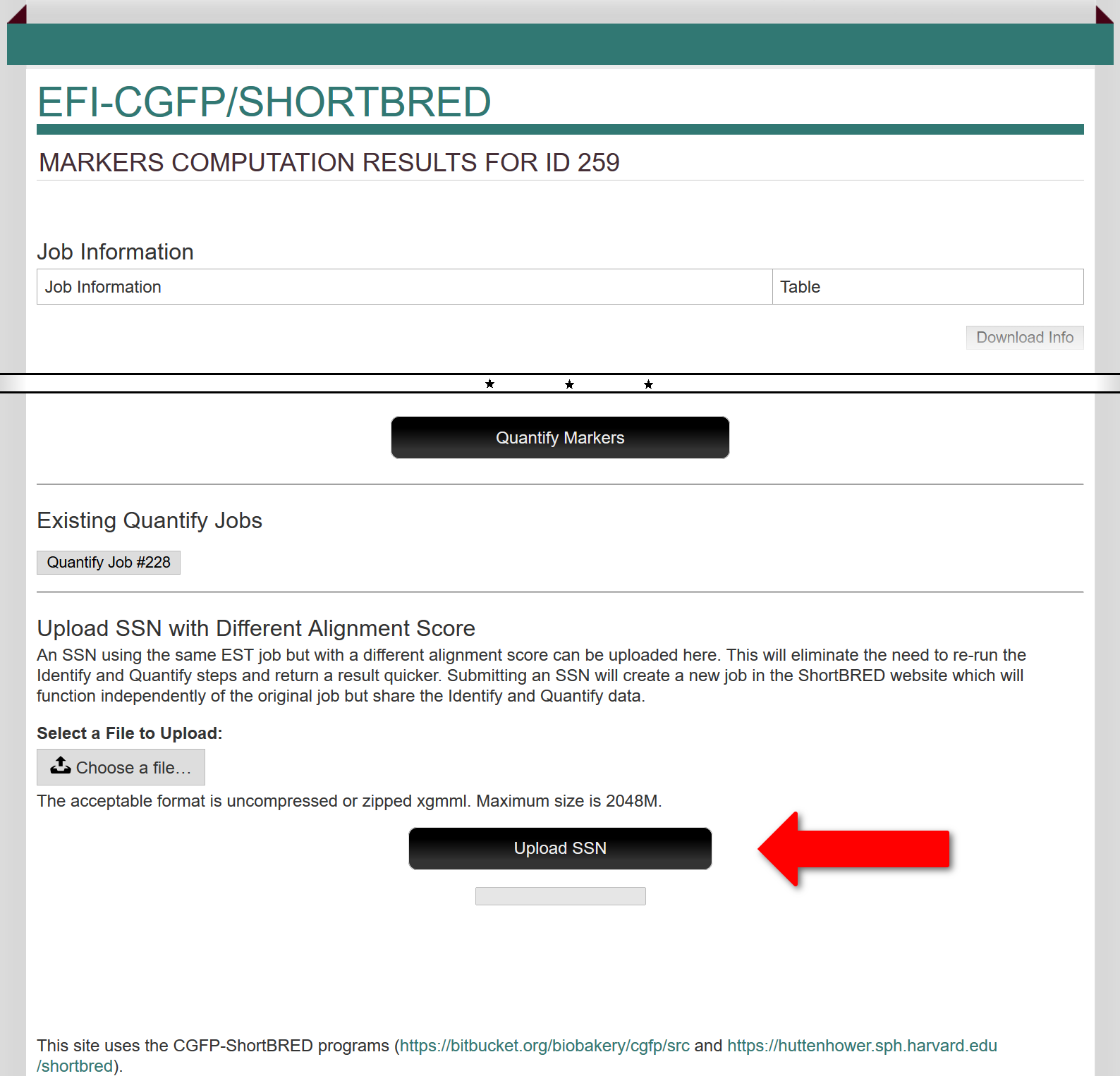
In addition, after both the Identify and Quantify steps have been completed for an input SSN, this page allows the user to upload a different Colored SSN ("child" SSN) and map the protein/cluster abundances to the new SSN (using the "Upload SSN with Different Alignment Score" section; red arrow). The new SSN must have been generated using the same EFI-EST job as the original SSN uploaded to EFI-CGFP and segregated with a different alignment score and/or a subset of the clusters. This upload eliminates the need to re-run the Identify and Quantify steps so the results will be obtained in a much shorter period of time (minutes instead of days!). Submitting an SSN will create a new job in the "Previous Jobs" panel (indicated as "Child" of the initial marker identification/metagenome quantification job) which functions independently of the original job but shares the Identify and Quantify data. The results for the "Child" job will include heat maps for the clusters and singletons in the uploaded SSN as well as the SSN with marker and metagenome hit node attributes (next section).
Click here to contact us for help, reporting issues, or suggestions.


