EFI - Genome Neighborhood Tool
EFI-GNT Input and Output Pages
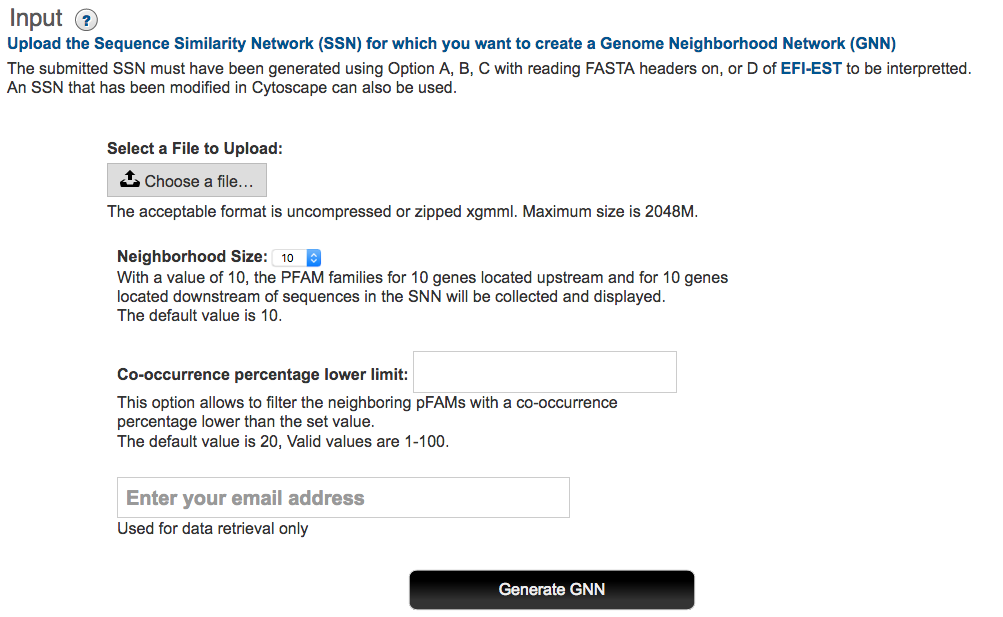
Start Page/Input
Acceptable SSNs are generated for an entire Pfam and/or InterPro protein family (from Option B of EFI-EST), a focused region of a family (from Option A of EFI-EST), a set of protein sequence that can be identified from FASTA headers (from option C of EFI-EST with header reading) or a list of recognizable UniProt and/or NCBI IDs (from option D of EFI-EST). An SSN manually modified within Cytoscape that originated from any of acceptable EFI-EST Options is also acceptable. SSNs that have been colored using the "Color SSN Utility" of EFI-EST and that originated from any of the available options are also acceptable.
SSNs generated with Option C of the EFI-EST without the header reading option selected will not work—the process for generating the GNN requires that the sequences have UniProt IDs.
The maximum size of the xgmml file is 2048 MB. The SSN may be either a full SSN (a node for each sequence) or a representative-node (rep-node) SSN (sequences sharing greater than a user-selected sequence identity are located in the same metanode). The xgmml file may be either uncompressed or zipped.
EFI-GNT uses a default ± 10 orf window to collect the genome neighbors; the user can select a smaller window (from ± 3 – ± 20 orfs) in the "Neighborhood Size" pull-down menu.
EFI-GNT collects all genome neighbors within the specified window. However, it will display a spoke node only if the query-neighbor co-occurrence frequency is greater than a specified value. The default value is 20%. A smaller value, e.g., 5%, should be used to find neighbors that co-occur with low frequency, often as the result of phylogenetically diverse genome arrangements of functionally linked pathway enzymes. As the co-occurrence frequency is decreased, a larger number of neighbors and Pfam families will be reported in the GNN and the signal-to-noise ratio will decrease.
As with EFI-EST, the user also inputs an e-mail address to which an e-mail containing a link to the results will be sent.
Download Page/Output
When the results are available (typically a few minutes, although the time required for the analysis increases with the number of query sequences in the input SSN), an e-mail with a link to the output is sent to the address provided by the user on the Start page. The link will be active for seven days.
A summary of the submission is available.
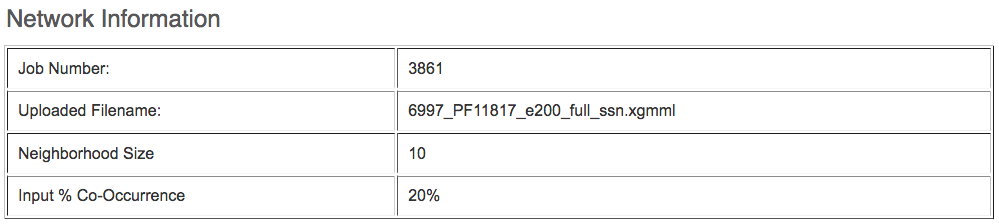
The EFI-GNT output is three xgmml files and several text/spreadsheet files.

Colored SSN: The colored version of the SSN described in the previous section is available for download as an xgmml file for viewing in Cytoscape. This SSN allows the user to quickly associate SSN cluster spoke nodes in the GNNs with clusters in the input query SSN.
Two formats of the GNN: The two formats of the GNN described in the previous section are available for download as xgmml files and viewing/analysis in Cytoscape:
- A cluster is present for each Pfam family (hub-node) that was identified as a neighbor to queries in the SSN clusters (spoke-nodes). This format allows the user to assess whether queries in multiple SSN clusters are neighbors to members of the same Pfam family and, therefore, may have the same in vitro activities and in vivo metabolic functions.
- A cluster is present for each query SSN cluster (hub-node) that was used to identify genome neighbors (spoke-nodes). This format allows the user to identify functionally linked enzymes, as deduced from genome proximity, that constitute the metabolic pathway in which the sequences in the query SSN cluster participate.
In addition to the colored SSN and GNNs, several text files/folders of text files are available for download:
- "UniProt ID-Color-Cluster Number Mapping Table" is a tab-delimited text file with three columns with headers, "UniProt ID", "Cluster Number", and "Cluster Color"; a description of this file and its use is provided in the Color SSN Utility section of the EFI-EST tutorial.
- "UniProt IDs per Cluster" is a folder of files for each cluster that list the UniProt IDs for the sequences in the cluster; a description of the files in this folder and their use was provided in the Color SSN Utility section of the EFI-EST tutorial.
- "FASTA Files per Cluster" is a folder of files for each cluster that contain the FASTA-formatted sequences for the sequences in the cluster; a description of the files in this folder and their use was provided in the Color SSN Utility section of the EFI-EST tutorial.
- "Pfam Neighbor Mapping Tables" is a folder of tab-delimited text files for each neighbor Pfam family that includes the following columns: "Query ID", "Neighbor ID", "Neighbor Pfam", "SSN Query Cluster #", "SSN Query Cluster Color", "Query-Neighbor Distance" (absolute value of distances in ORFs), and "Query-Neighbor Directions" (relative directions of transcription). These files can be used with the Cytoscape BridgeDB App to add these columns as node attributes to the SSN for the neighbor Pfam family so that the neighbors can be located/analyzed in the sequence-function context of the family. The "SSN Query Cluster Color" node attributes can be used with "pass-through mapping" in the Cytoscape Style Color panel to color the nodes in the neighbor Pfam family with the colors in the colored SSN generated by EFI-GNT 1.0, thereby facilitating the determination of whether the neighbors identified in the genome neighborhoods are orthologous. A concatenated file that contains all of the information is also available and may be more convenient for adding node attributes to SSNs for multiple Pfam families.
- "Neighbors without Pfam assigned per Cluster" is a folder of tab-delimited text files for each SSN cluster that lists the accession IDs of neighbors not assigned to Pfam families. These files allow SSNs to be generated with Option D of EFI-EST 2.0 so that protein families not curated by Pfam can be identified. The user should use an alignment score of ~20 to filter the SSN; in most cases, this alignment score will segregate the SSN into protein families.
- "No Matches/No Neighbors File" is a tab-delimited text file with two columns: "UniProt ID" and "No Match/No Neighbor" ("nomatch" or "neighbor"). The same information is included in the colored SSN generated by EFI-GNT 1.0 (vide infra).
Click here to contact us for help, reporting issues, or suggestions.


