EFI - Genome Neighborhood Tool
EFI-GNT allows exploration of the genome neighborhoods for sequence similarity network (SSN) clusters in order to facilitate the assignment of function within protein families and superfamilies.
In GNT Submission, each sequence within a SSN is used as a query for interrogation of its genome neighborhood. A colored SSN identifying clusters, Genome Neighborhood Networks (GNNs) providing statistical analysis of neighboring Pfam families, Genome Neighborhood Diagrams (GNDs), sets of IDs and sequences per cluster and additional files are created. For the Retrieve Neighorhood Diagrams option, only GNDs will be created.
A listing of new features and other information pertaining to GNT is available on the release notes page.
The GNT database uses UniProt 2024_01, and ENA downloaded on January 2024.
In a submitted SSN, each sequence is considered as a query. Information associated with protein encoding genes that are neighbors of input queries (within a defined window on either side) are collected from sequence files for bacterial (prokaryotic and archaeal) and fungal genomes in the European Nucleotide Archive (ENA) database. The neighboring genes are sorted into neighbor Pfam families. For each cluster, the co-occurrence frequencies of the identified neighboring Pfam families with the input queries are calculated.
The provided sequence is used as the query for a BLAST search of the UniProt database. The retrieved sequences are used to generate genomic neighborhood diagrams.
If the Sequence Database is set to UniRef90, the resulting GNDs will also include GNDs for UniRef90 cluster IDs that group together UniProt sequences by 90% sequence identiy. For UniRef50, the GNDs will also include UniProt sequences that are grouped by 50% sequence identity. Any of the "Exclude Fragments" options will exclude UniProt-defined sequence fragments.
The genomic neighborhoods are retreived for the UniProt, NCBI, EMBL-EBI ENA, and PDB identifiers that are provided in the input box below. Not all identifiers may exist in the EFI-GNT database so the results will only include diagrams for sequences that were identified.
If the Sequence Database is set to UniRef90, the resulting GNDs will also include GNDs for UniRef90 cluster IDs that group together UniProt sequences by 90% sequence identiy. For UniRef50, the GNDs will also include UniProt sequences that are grouped by 50% sequence identity. Any of the "Exclude Fragments" options will exclude UniProt-defined sequence fragments.
The genomic neighborhoods are retreived for the UniProt, NCBI, EMBL-EBI ENA, and PDB identifiers that are identified in the FASTA headers. Not all identifiers may exist in the EFI-GNT database so the results will only include diagrams for sequences that were identified.
EFI-Genome Neighborhood Tool Overview
The EFI-GNT (EFI Genome Neighborhood Tool) is focused on placing protein families and superfamilies into a genomic context. A sequence similarity network (SSN) is used as an input. Each sequence within a SSN is used as a query for interrogation of its genome neighborhood.
EFI-GNT enables exploration of the genome neighborhoods for sequences in SSN clusters in order to facilitate their assignment of function.
EFI-GNT Acceptable Input
EFI-GNT is compatible with SSN generated by the EFI-Enzyme Similarity Tool (EFI-EST). Acceptable SSNs are generated for an entire Pfam and/or InterPro protein family (EFI-EST option B), a focused region of a family (option A), a set of protein sequence that can be identified from FASTA headers (from option C with “Header Reading” activated) or a list of recognizable UniProt and/or NCBI IDs (from option D). SSNs manually modified within Cytoscape are accepted. SSNs that have been colored using the "Color SSN Utility" are also accepted. SSNs generated from FASTA sequences (option C) without the "Read Header" option activated are not accepted.
Principle of GNT Analysis
EFI-GNT provides statistical analysis, per SSN cluster, of genome context for bacterial, archeal and fungal sequences, in order to identify possible functional linkage. Sequences from the SSN analyzed are used as query for retrieval of their genome neighborhood. The user specifies the neighborhood size (±N orfs from the SSN query) and minimum query-neighbor co-occurrence frequency for the outputs.
EFI-GNT Output
EFI-GNT identifies each SSN cluster and assigns it a unique color. A colored SSN is produced. It then interrogates the European Nucleotide Archive (ENA; https://www.ebi.ac.uk/ena) to obtain the genome contexts of each sequence, sorts neighbors into Pfam families, and provides three specific outputs. Firstly, a GNN network in which each SSN cluster is a hub node with its spoke nodes identified neighboring Pfam families (for identifying candidates for pathway enzymes); secondly, a GNN network in which each neighbor Pfam family is a hub node with its spoke nodes that SSN clusters that identify this Pfam as a neighbor (for identifying divergent clusters that are orthologues); and thirdly, genome neighborhood diagrams (GNDs) for visual representations of the neighborhoods for the sequences in each SSN cluster (for visual inspection of synteny and the presence/absence of functionally linked proteins).
Direct Genomic Neighborhood Diagrams (GND) Generation
The "Retrieve neighborhood diagrams" allows exploring of neighboring genes for specific queries. You can submit a single sequence that is used as the query for a BLAST search of the UniProt database. The retrieved sequences are used to generate GNDs. GNDs can be generated from a provided list of IDs or even from FASTA sequences, by collecting IDs from FASTA headers.
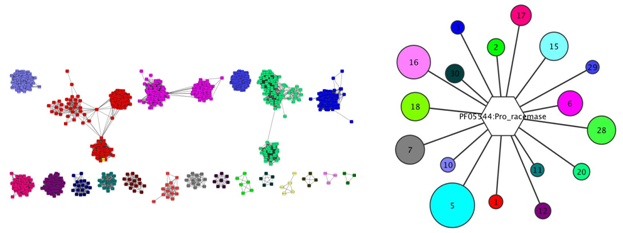
Figure 1: Examples of colored SSN (left) and a hub-and-spoke cluster from a GNN (right).
Recommended Reading
Rémi Zallot, Nils Oberg, John A. Gerlt, "Democratized" genomic enzymology web tools for functional assignment, Current Opinion in Chemical Biology, Volume 47, 2018, Pages 77-85, https://doi.org/10.1016/j.cbpa.2018.09.009
John A. Gerlt, Genomic enzymology: Web tools for leveraging protein family sequence–function space and genome context to discover novel functions, Biochemistry, 2017 - ACS Publications
UniProt Version: 2024_01
InterPro Version: 98
ENA Version: January 2024
Click here to contact us for help, reporting issues, or suggestions.


