Enzyme Function Initiative Tools
This website contains a collection of webtools for creating and interacting with sequence similarity networks (SSNs) and genome neighborhood networks (GNNs). These tools originated in the Enzyme Function Initiative, a NIH-funded research project to develop a sequence / structure-based strategy for facilitating discovery of in vitro enzymatic and in vivo metabolic / physiological functions of unknown enzymes discovered in genome projects.
The Enzyme Function Initiative tools are hosted at the Carl R. Woese Institute for Genomic Biology (IGB), at the University of Illinois at Urbana-Champaign. The development of these tools is headed by John A. Gerlt (PI), aided by scientist Rémi Zallot and software engineers Dan Davidson, David Slater, Russell Davidson, and Nils Oberg. The computational infrastracture is supported by the Computer and Network Resource Group at IGB.
An article in the 2023 Journal of Molecular Biology Computation Resources for Molecular Biology was recently published and is available on the training resources page.
A "From The Bench" article was published in Biochemistry in 2019 and is available on the training resources page.
The web tools have been described in five publications:
Nils Oberg, Rémi Zallot, and John A. Gerlt, EFI-EST, EFI-GNT, and EFI-CGFP: Enzyme Function Initiative (EFI) Web Resource for Genomic Enzymology Tools. J Mol Biol 2023. https://doi.org/10.1016/j.jmb.2023.168018
Rémi Zallot, Nils Oberg, and John A. Gerlt, The EFI Web Resource for Genomic Enzymology Tools: Leveraging Protein, Genome, and Metagenome Databases to Discover Novel Enzymes and Metabolic Pathways. Biochemistry 2019 58 (41), 4169-4182. https://doi.org/10.1021/acs.biochem.9b00735
Rémi Zallot, Nils Oberg, John A. Gerlt, "Democratized" genomic enzymology web tools for functional assignment, Current Opinion in Chemical Biology, Volume 47, 2018, Pages 77-85, https://doi.org/10.1016/j.cbpa.2018.09.009
John A. Gerlt, Genomic enzymology: Web tools for leveraging protein family sequence–function space and genome context to discover novel functions, Biochemistry. Volume 56, 2017, Pages 4293-4308. https://doi.org/10.1021/acs.biochem.7b00614
John A. Gerlt, Jason T. Bouvier, Daniel B. Davidson, Heidi J. Imker, Boris Sadkhin, David R. Slater, Katie L. Whalen, Enzyme Function Initiative-Enzyme Similarity Tool (EFI-EST): A web tool for generating protein sequence similarity networks, Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics, Volume 1854, Issue 8, 2015, Pages 1019-1037, ISSN 1570-9639, https://dx.doi.org/10.1016/j.bbapap.2015.04.015
These webtools use NCBI BLAST and CD-HIT to create SSNs and GNNs. The computationally-guided functional profiling tool uses the CGFP programs from the Balskus Lab (https://bitbucket.org/biobakery/cgfp/src) and ShortBRED from the Huttenhower Lab (http://huttenhower.sph.harvard.edu/shortbred). The data used originate from the UniProt Consortium databases and the InterPro and ENA databases from EMBL-EBI.
Click here to contact us for help, reporting issues, or suggestions.








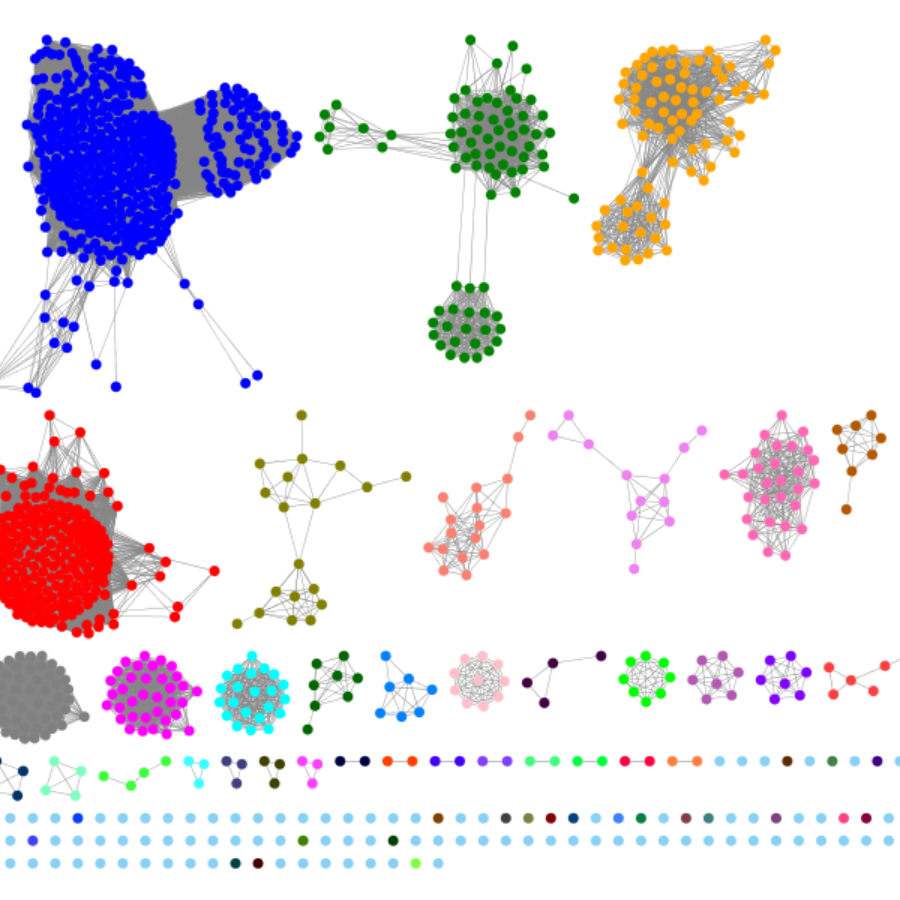 SSN creation from sequences, Pfam or InterPro famil(ies), and
UniProt and/or NCBI protein accession IDs.
SSN creation from sequences, Pfam or InterPro famil(ies), and
UniProt and/or NCBI protein accession IDs.
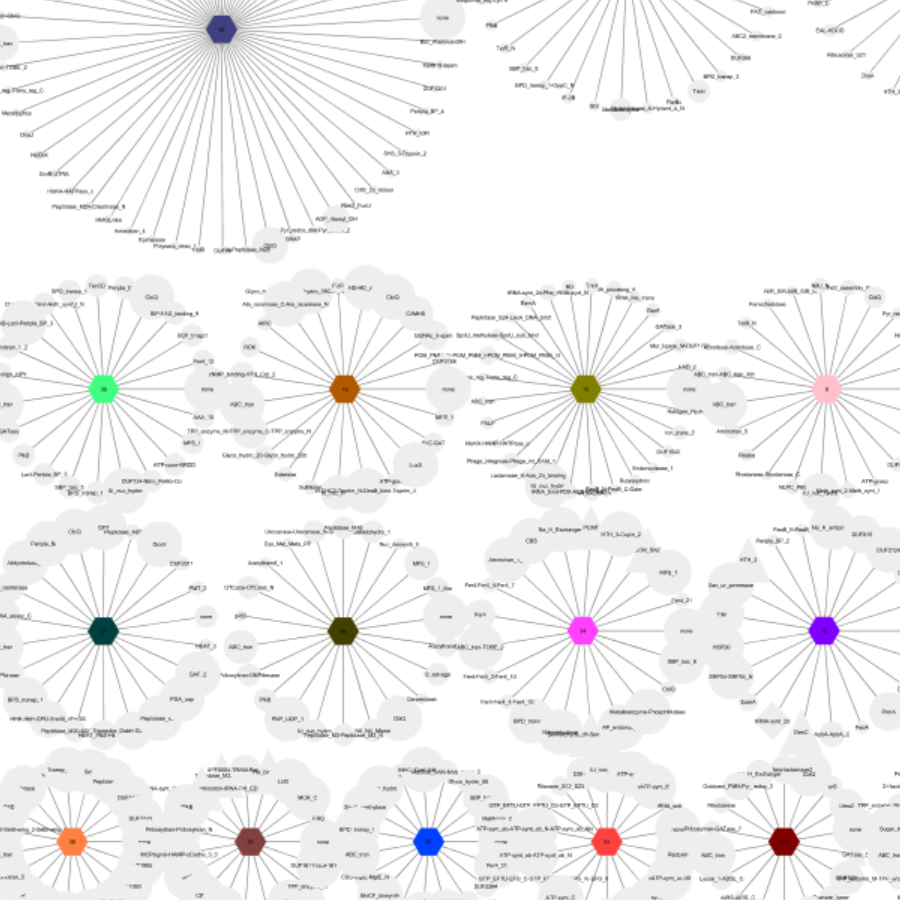 GNN creation from sequence similarity networks.
GNN creation from sequence similarity networks.
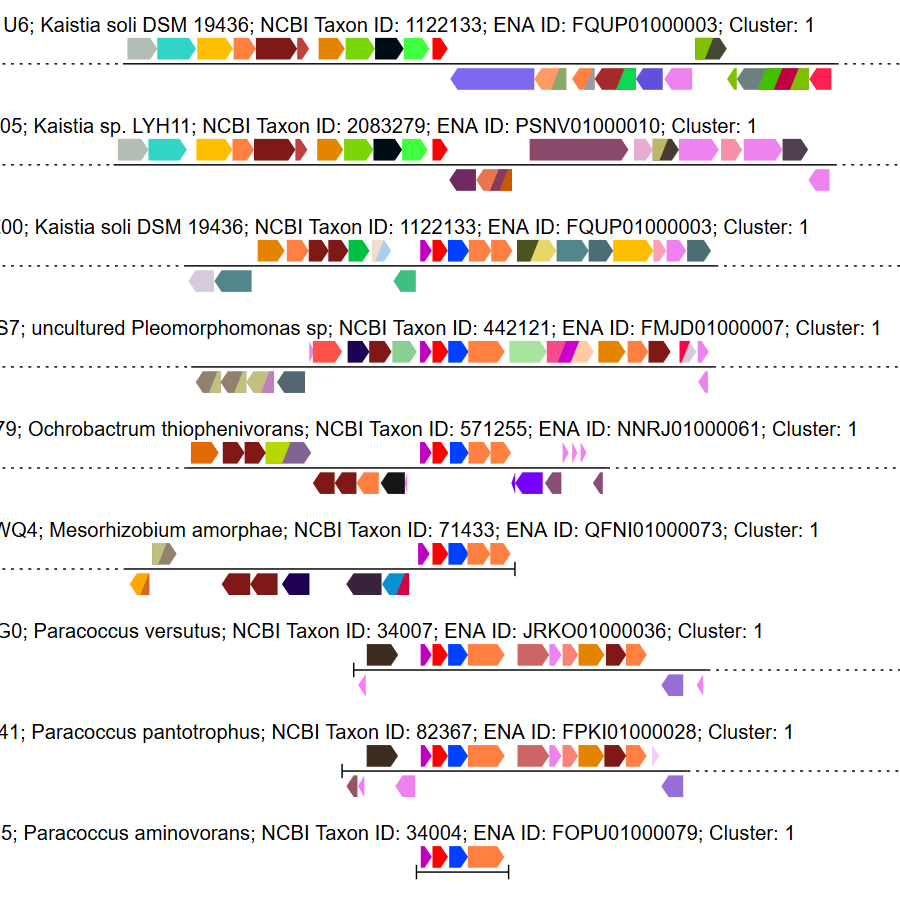 Genome neighborhood diagrams from sequences, sequence IDs, or GNNs.
Genome neighborhood diagrams from sequences, sequence IDs, or GNNs.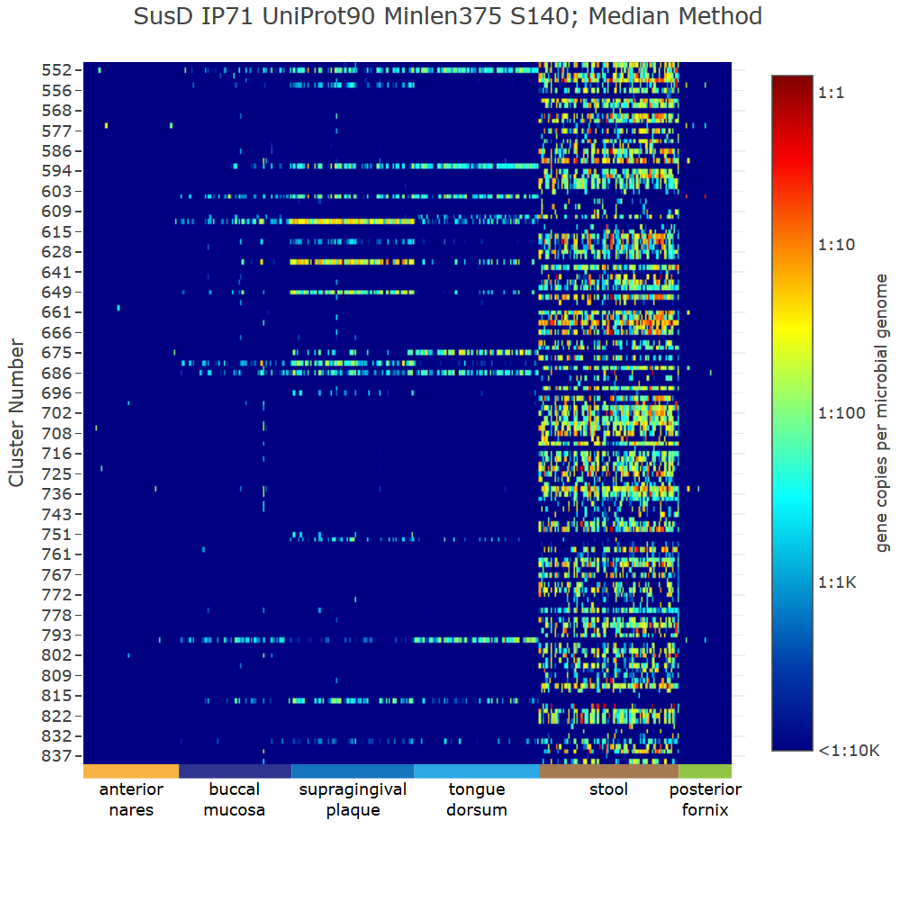 Chemically-guided functional profiling using ShortBRED and the CGFP tools from
the Balskus and Huttenhower Labs at Harvard University.
Chemically-guided functional profiling using ShortBRED and the CGFP tools from
the Balskus and Huttenhower Labs at Harvard University.
